What's New
International Sheep Genomics Consortium

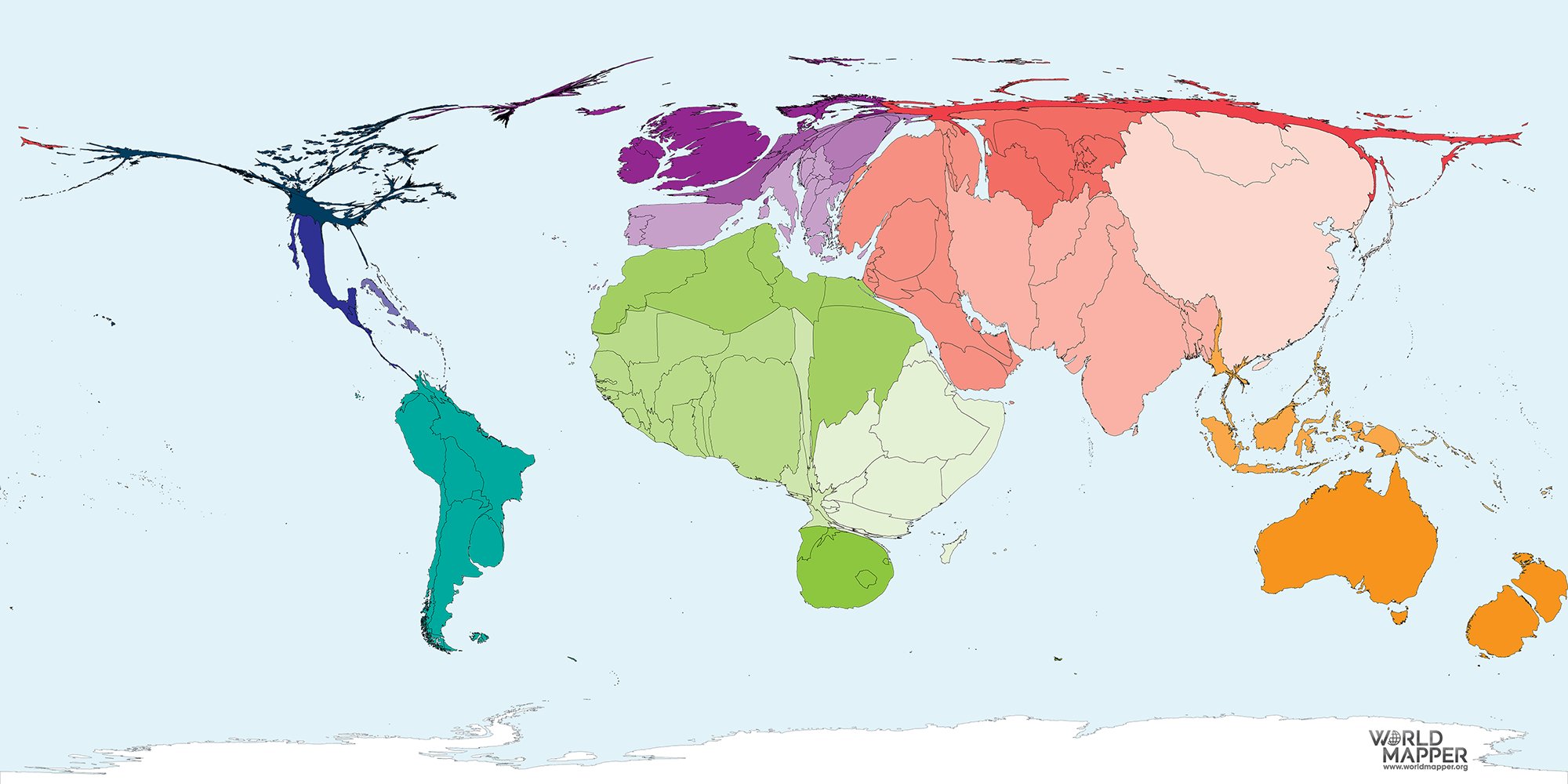

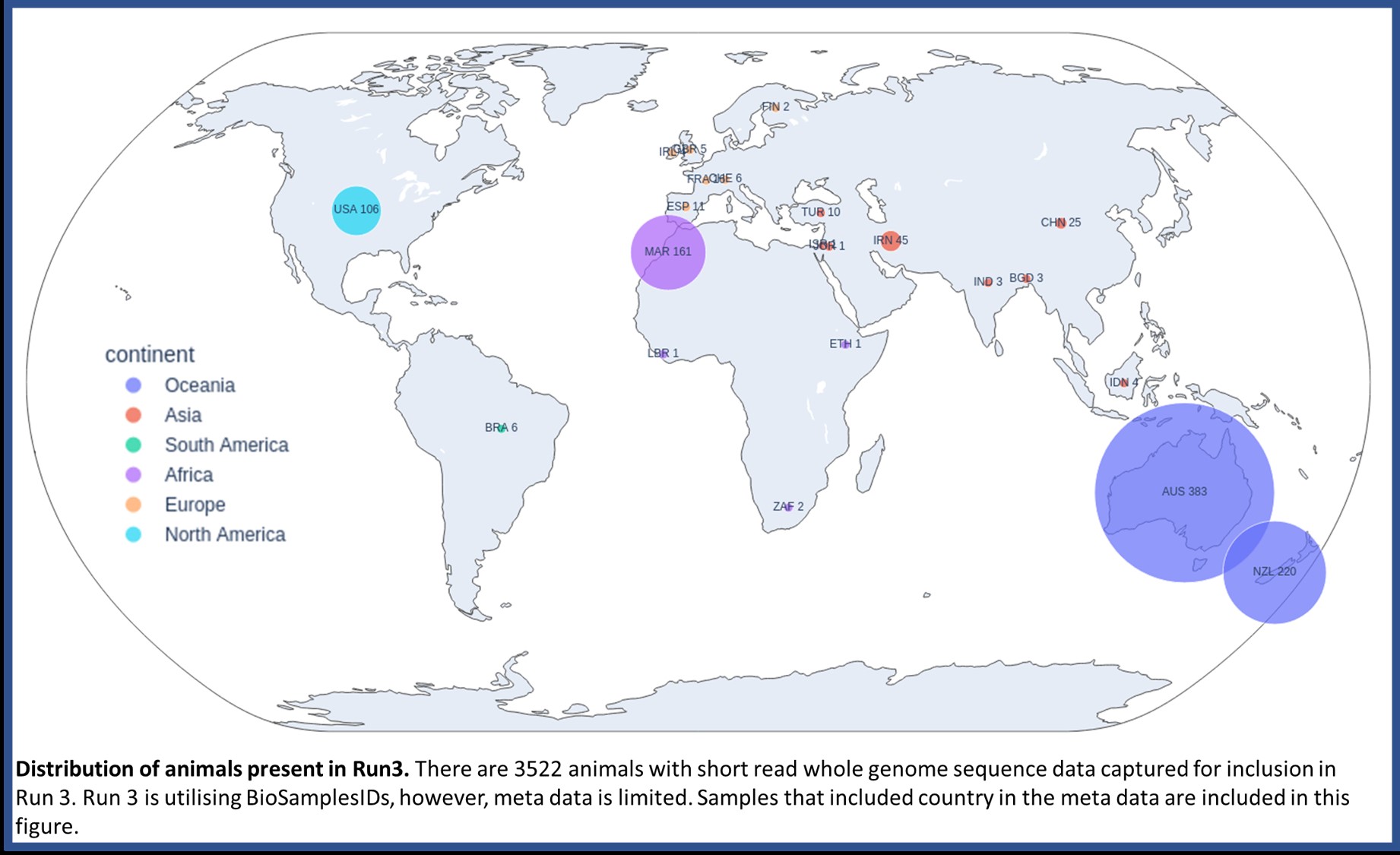
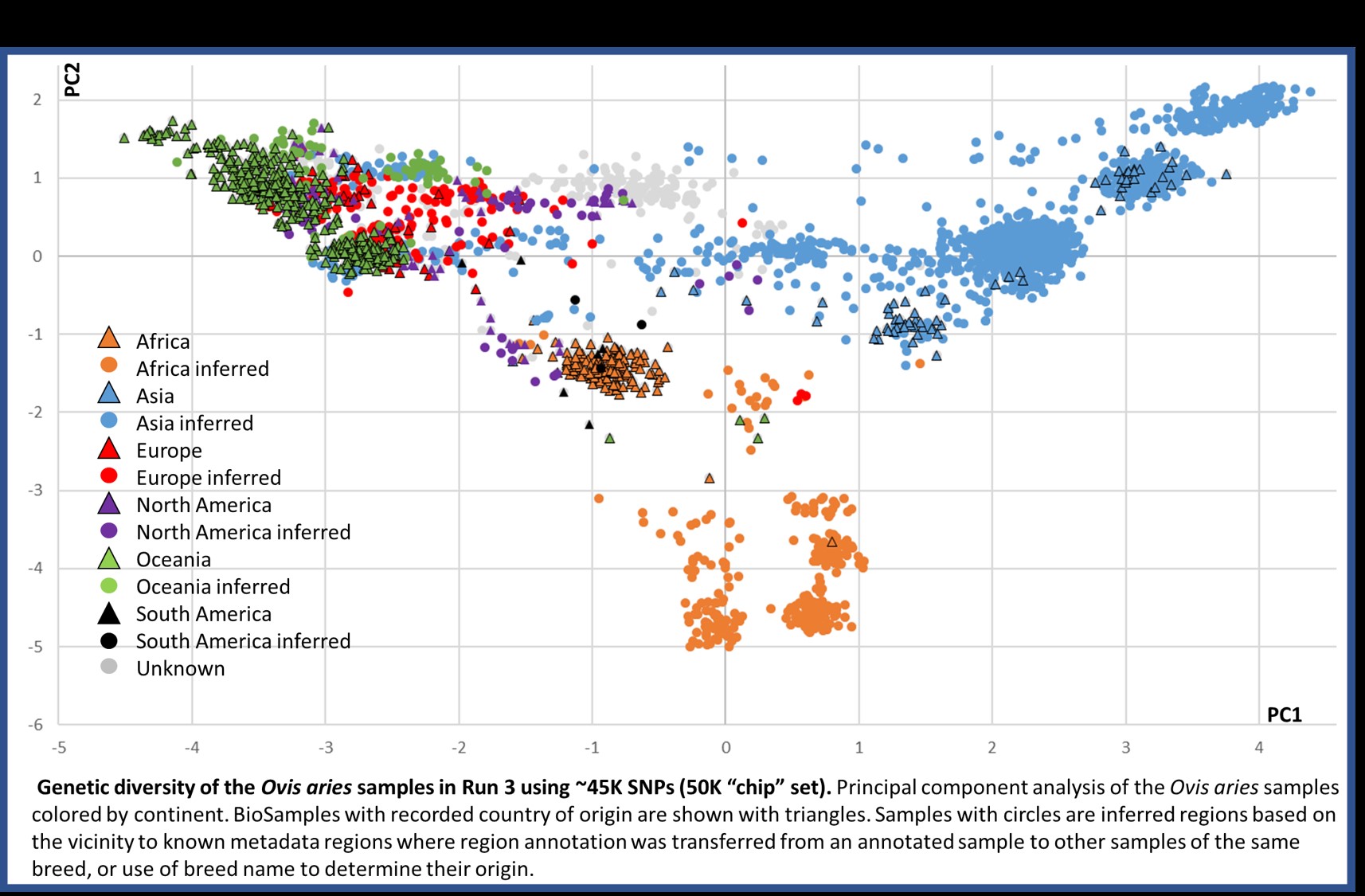
Announcing Jan 2025: 3522 genomes have been included in Run3 of the 1000 genomes project

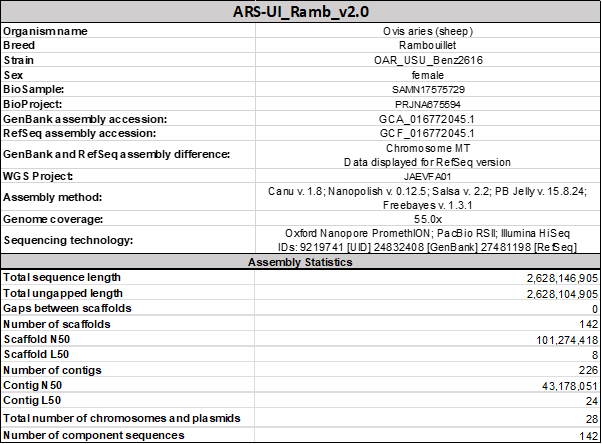
Completed ARS-UI_Ramb_v2.0 genome assembly and annotation
ARS-UI_Ramb_v2.0 .assembly and annotation has now been formally published in Feb 2022. Davenport et al. An improved ovine reference genome assembly to facilitate in-depth functional annotation of the sheep genome GigaScience, Volume 11, 2022, giab096 link
Details of this annotation, including statistics on the annotation products, the input data used in the pipeline and intermediate alignment results, can also be found in Annotation Release 104 of ARS-UI_Ramb_v2.0 link
Background to ISGC
The International Sheep Genomics Consortium (ISGC) is a partnership of scientists and funding agencies from Australia, Austria, Brazil, China, Finland, France, Germany, Greece, India, Iran, Israel, Italy, Kenya, New Zealand, Norway, Saudi Arabia, Spain, Switzerland, Turkey, United Kingdom and United States to develop public genomic resources that will help researchers find genes associated with production, quality and disease traits in sheep.
The project commenced informally in 2002 with the creation of a high quality ovine BAC library, and was built on an existing collaboration for the International Mapping Flock that was created nearly a decade earlier.
This work has continued and is most well known for the initial sequencing of the sheep genome and the creation of several SNP chip arrays: specifically the publicly available Illumina 50K and the Illumina 15K SNP chips. The ISGC was also involved in the creation of the Illumina HD 600K chip which is available upon request (see contacts). However, its major ongoing function has been sequencing and annotation of the sheep genome. This includes projects such as FAANG and the SheepGenomesDB commonly called the 1000 genomes sheep project.
Sheep Genome Assemblies
Please be aware that the various sheep genome assemblies are labelled differently in the different repositories. This has significant implications when identifying SNPs and other features in published papers. The initial assembly Oar_v1.0 was used to build the 50K chip and is still available at UCSC labelled as ISGC Ovis_aries_1.0. However, the four assemblies listed below are those that most published work has utilised.
Oar_v3.1 This version was published in Science in 2014 from the sequence of a Texel animal. The fully annotated genome is available via ENSEMBL, NCBI and UCSC.
Oar_v4.0 In 2015 the ISGC released Oar_v4.0 whereby long read technology (PacBio RSII) was utilised to improve the Oar_v3.1 assembly.
Oar_rambouillet_v1.0 In 2017 Baylor College of Medicine Human Genome Sequencing Center released a genome assembly from the Ramboullet breed. The genome assembly utilised a combination of Illumina short reads and PacBio RSII long reads.
ARS-UI_Ramb_v2.0 This is an improved genome assembly for OAR_USU_Benz2616 submitted by University of Idaho. Davenport KM, Bickhart DM, Worley K, Murali SC, Salavati M, Clark EL, Cockett NE, Heaton MP, Smith TP, Murdoch BM, Rosen BD. An improved ovine reference genome assembly to facilitate in-depth functional annotation of the sheep genome. GigaScience. 2022 Feb 4;11.
In addition, annotation of Rambouillet (OAR_USU_Benz2616) genome is underway via The Ovine FAANG project, led by Brenda Murdoch University of Idaho and is supported by the National Institute of Food and Agriculture, U.S. Department of Agriculture, award number USDA-NIFA-2017-67016-26301.
Variant detection for user-supplied genome sequences
Results from Run2
Results from Run3
ISGC SNP chip array genome positions
The SNPs on the consortium arrays (Illumina 15k, 50k and HD chips) have been mapped to ARS-UI_Ramb_v2.0. Probe sequences were taken from the Illumina manifests and mapped onto the Rambouillet genome (GCA_016772045.1) using bwa mem v0.7.17-r1188 with default settings (Indels were ignored). For each SNP a probe pair was constructed by using AlleleA_ProbeSeq and appending either the reference or the alternative allele. Only probe pairs were accepted that passed following filters.
The arrays were in addition mapped to Oar_v3.1, Oar_v4 and Oar_rambouillet_v1.0 to enable comparison of mapping approach to NCBI and Ensembl. SNP name, position and allele from the consortium arrays available on Figshare
